-Search query
-Search result
Showing all 47 items for (author: falk & s)

EMDB-40094: 
17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer
Method: single particle / : Kwon HJ, Zhang J, Kosikova M, Tang WC, Rodriguez UO, Peng HQ, Meseda CA, Pedro CL, Schmeisser F, Lu JM, Zhou B, Davis CT, Wentworth DE, Chen WH, Shriver MC, Pasetti MF, Weir JP, Chen B, Xie H

EMDB-40095: 
17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer
Method: single particle / : Kwon HJ, Zhang J, Kosikova M, Tang WC, Rodriguez UO, Peng HQ, Meseda CA, Pedro CL, Schmeisser F, Lu JM, Zhou B, Davis CT, Wentworth DE, Chen WH, Shriver MC, Pasetti MF, Weir JP, Chen B, Xie H

PDB-8gjm: 
17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer
Method: single particle / : Kwon HJ, Zhang J, Kosikova M, Tang WC, Rodriguez UO, Peng HQ, Meseda CA, Pedro CL, Schmeisser F, Lu JM, Zhou B, Davis CT, Wentworth DE, Chen WH, Shriver MC, Pasetti MF, Weir JP, Chen B, Xie H

PDB-8gjn: 
17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer
Method: single particle / : Kwon HJ, Zhang J, Kosikova M, Tang WC, Rodriguez UO, Peng HQ, Meseda CA, Pedro CL, Schmeisser F, Lu JM, Zhou B, Davis CT, Wentworth DE, Chen WH, Shriver MC, Pasetti MF, Weir JP, Chen B, Xie H

EMDB-14619: 
Cryo-EM structure of POLRMT mutant.
Method: single particle / : Das H, Hallberg BM

PDB-7zc4: 
Cryo-EM structure of POLRMT mutant.
Method: single particle / : Das H, Hallberg BM

EMDB-13736: 
Cryo-EM structure of POLRMT in free form.
Method: single particle / : Das H, Hallberg BM

EMDB-13796: 
2021-10-18
Method: single particle / : Das H, Hallberg BM

PDB-7pzr: 
Cryo-EM structure of POLRMT in free form.
Method: single particle / : Das H, Hallberg BM

EMDB-13735: 
Mitochondrial DNA dependent RNA polymerase homodimer.
Method: single particle / : Das H, Hallberg BM

PDB-7pzp: 
Mitochondrial DNA dependent RNA polymerase homodimer.
Method: single particle / : Das H, Hallberg BM

EMDB-25397: 
CSDaV GFP mutant
Method: single particle / : Guo F, Matsumura EE, Falk BW

EMDB-25398: 
CSDaV wild-type
Method: single particle / : Guo F, Matsumura EE, Falk BW

PDB-7sqy: 
CSDaV GFP mutant
Method: single particle / : Guo F, Matsumura EE, Falk BW

PDB-7sqz: 
CSDaV wild-type
Method: single particle / : Guo F, Matsumura EE, Falk BW

EMDB-13146: 
Human mitochondrial Lon protease with substrate in the ATPase domain
Method: single particle / : Valentin Gese G, Shahzad S, Hallberg BM

EMDB-13147: 
Human mitochondrial Lon protease without substrate
Method: single particle / : Valentin Gese G, Shahzad S, Hallberg BM

EMDB-13148: 
Human mitochondrial Lon protease with substrate in the ATPase and protease domains
Method: single particle / : Valentin Gese G, Shahzad S, Hallberg BM

EMDB-23791: 
Subtomogram average of Photosystem II 2D semicrystalline array on thylakoid membranes isolated from Phaeodactylum tricornutum
Method: subtomogram averaging / : Jiang J, Cheong KY, Falkowski PG, Dai W

EMDB-11162: 
Amyloid fibril morphology i (in vitro) from murine SAA1.1 protein
Method: helical / : Bansal A, Schmidt M, Faendrich M

EMDB-11163: 
Amyloid fibril morphology ii (in vitro) from murine SAA1.1 protein
Method: helical / : Bansal A, Schmidt M, Fandrich M

EMDB-11164: 
Amyloid fibril morphology II (ex vivo) from murine SAA1.1 protein.
Method: helical / : Bansal A, Schmidt M, Faendrich M

PDB-6zcf: 
Amyloid fibril morphology i (in vitro) from murine SAA1.1 protein
Method: helical / : Bansal A, Schmidt M, Faendrich M

PDB-6zcg: 
Amyloid fibril morphology ii (in vitro) from murine SAA1.1 protein
Method: helical / : Bansal A, Schmidt M, Fandrich M

PDB-6zch: 
Amyloid fibril morphology II (ex vivo) from murine SAA1.1 protein.
Method: helical / : Bansal A, Schmidt M, Faendrich M

EMDB-11679: 
Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor.
Method: single particle / : Hillen HS, Bonekamp N, Peter B, Felser A, Bergbrede T, Choidas A, Horn M, Unger A, di Lucrezia R, Atanassov I, Li X, Koch U, Menninger S, Boros J, Habenberger P, Giavalisco P, Cramer P, Denzel M, Nussbaumer P, Klebl B, Falkenberg M, Gustafsson CM, Larsson NG

PDB-7a8p: 
Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor.
Method: single particle / : Hillen HS, Bonekamp N, Peter B, Felser A, Bergbrede T, Choidas A, Horn M, Unger A, di Lucrezia R, Atanassov I, Li X, Koch U, Menninger S, Boros J, Habenberger P, Giavalisco P, Cramer P, Denzel M, Nussbaumer P, Klebl B, Falkenberg M, Gustafsson CM, Larsson NG

EMDB-0539: 
Averaged structure of Photosystem II holocomplex on thylakoid membrane from diatom
Method: subtomogram averaging / : Chen M, Kuang X, Levitan O, Falkowski P, Dai W

EMDB-0540: 
Averaged structure of Photosystem II 2D crystalline array on thylakoid membrane from diatom
Method: subtomogram averaging / : Chen M, Kuang X, Levitan O, Falkowski P, Dai W

EMDB-8910: 
Cryo-EM structure of murine AA amyloid fibril
Method: helical / : Liberta F, Fandrich M, Schmidt M

EMDB-9232: 
Cryo-EM structure of human AA amyloid fibril
Method: helical / : Rennegarbe M, Liberta F, Fandrich M, Schmidt M

PDB-6dso: 
Cryo-EM structure of murine AA amyloid fibril
Method: helical / : Loerch S, Liberta F, Grigorieff N, Fandrich M, Schmidt M

PDB-6mst: 
Cryo-EM structure of human AA amyloid fibril
Method: helical / : Loerch S, Rennegarbe M, Liberta F, Grigorieff N, Fandrich M, Schmidt M

EMDB-0128: 
Human nuclear RNA exosome EXO-10-MPP6 complex
Method: single particle / : Gerlach P, Schuller JM, Falk S, Basquin J, Conti E

PDB-6h25: 
Human nuclear RNA exosome EXO-10-MPP6 complex
Method: single particle / : Gerlach P, Schuller JM, Falk S, Basquin J, Conti E

EMDB-0127: 
Human nuclear RNA exosome EXO-14 complex
Method: single particle / : Gerlach P, Schuller JM, Falk S, Basquin J, Conti E

EMDB-4302: 
Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors
Method: single particle / : Schuller JM, Falk S, Conti E

PDB-6ft6: 
Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors
Method: single particle / : Schuller JM, Falk S, Conti E

EMDB-4301: 
Structure of the nuclear RNA exosome
Method: single particle / : Schuller JM, Falk S, Conti E

PDB-6fsz: 
Structure of the nuclear RNA exosome
Method: single particle / : Schuller JM, Falk S, Conti E

EMDB-5918: 
Negative stain reconstruction of PGT151 Fab in complex with BG505 Env with transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB

EMDB-5919: 
Negative stain reconstruction of PGT151 Fab in complex with JR-FL Env with transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB
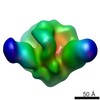
EMDB-5920: 
Negative stain reconstruction of PGT151 Fab in complex with cleaved IAVI C22 Env trimer containing the transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB
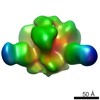
EMDB-5921: 
Negative stain reconstruction of PGT151 Fab in complex with the soluble Env trimer BG505 SOSIP
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB

EMDB-5409: 
A 3-D cryo-electron structure of bacteriophage phi92 contractile tail
Method: single particle / : Nazarov S, Bowman VD, Leiman PG

EMDB-2063: 
A 3-D cryo-electron microscopy structure of bacteriophage phi92 capsid
Method: single particle / : Browning C, Nazarov S, Bowman V, Leiman P

EMDB-2064: 
A 3-D cryo-electron structure of bacteriophage phi92 baseplate
Method: single particle / : Browning C, Nazarov S, Bowman VD, Leiman PG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
